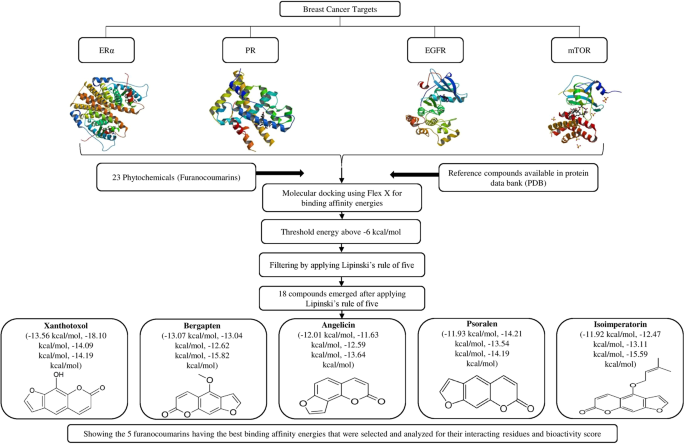
Structure Based Multitargeted Molecular Docking Analysis of Selected Furanocoumarins against Breast Cancer | Scientific Reports
![Values of binding affinity [kcal/mol] of the ligand molecule relative... | Download Scientific Diagram Values of binding affinity [kcal/mol] of the ligand molecule relative... | Download Scientific Diagram](https://www.researchgate.net/publication/334904554/figure/tbl3/AS:787624000241664@1564795603085/Values-of-binding-affinity-kcal-mol-of-the-ligand-molecule-relative-to-carbon-nanotubes.png)
Values of binding affinity [kcal/mol] of the ligand molecule relative... | Download Scientific Diagram

Relative binding affinity (ΔΔGXP = docking score (Glide XP in kcal/mol)... | Download Scientific Diagram
Is Kd (the dissociation constant) for a given protein-ligand pair directly correlated to binding affinity (kcal/mol, Rosetta Energy Units, etc.)? - Quora
Estimating the ligand‐binding affinity via λ‐dependent umbrella sampling simulations,Journal of Computational Chemistry - X-MOL

Values of binding affinity (kcal/mol) of Indol_4_9 molecule relative to... | Download Scientific Diagram

Binding Affinity Prediction for Protein–Ligand Complexes Based on β Contacts and B Factor | Journal of Chemical Information and Modeling
A computational binding affinity estimation protocol with maximum utilization of experimental data : A case study for Adenosine
![Values of binding affinity [kcal/mol] of ligand molecule relative to... | Download Scientific Diagram Values of binding affinity [kcal/mol] of ligand molecule relative to... | Download Scientific Diagram](https://www.researchgate.net/publication/334904554/figure/tbl2/AS:787624000249857@1564795603060/Values-of-binding-affinity-kcal-mol-of-ligand-molecule-relative-to-functionalized-C-60.png)
Values of binding affinity [kcal/mol] of ligand molecule relative to... | Download Scientific Diagram
AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility | PLOS Computational Biology

The average binding affinity (in kcal/mol) calculated using AutoDock... | Download Scientific Diagram
![Graphical presentation of the values of binding affinity [kcal/mol] of... | Download Scientific Diagram Graphical presentation of the values of binding affinity [kcal/mol] of... | Download Scientific Diagram](https://www.researchgate.net/publication/334214645/figure/fig2/AS:776775680393219@1562209162818/Graphical-presentation-of-the-values-of-binding-affinity-kcal-mol-of-the-ligand-CisPt.png)
Graphical presentation of the values of binding affinity [kcal/mol] of... | Download Scientific Diagram

A) Binding affinities (docking, kcal/mol), dissociation constants (Kd,... | Download Scientific Diagram

Binding affinity (kcal/mol) for different molecules for subunit F and... | Download Scientific Diagram
Applying Physics-Based Scoring to Calculate Free Energies of Binding for Single Amino Acid Mutations in Protein-Protein Complexes | PLOS ONE


![Values of binding affinity [kcal/mol] of Lig1 molecule relative to... | Download Scientific Diagram Values of binding affinity [kcal/mol] of Lig1 molecule relative to... | Download Scientific Diagram](https://www.researchgate.net/publication/334383636/figure/tbl1/AS:779299149844481@1562810804843/Values-of-binding-affinity-kcal-mol-of-Lig1-molecule-relative-to-nanosystems-obtained.png)





